Goals in Part 3
Continue the DNAToolkits
in part 1,with following functions added:
Transcription: transform DNA string to RNA string, which means replace 'T's with 'U'sDNA complementary strand : reverse & complement Use """{text}"""
to comment your function (so that you don't get confused when the scratch is too long).Colorize the nucleotide output Restructure and re-format the output.
1. Transcription
Very easy to implement.
def transcription(seq):
"""DNA -> RNA Transcription. Replacing Thymine with Uracil"""
return seq.replace("T", "U")复制
The use of function {string}.replace(a,b)
is quite obvious. Just to replace a
with b
in whatever {string} is.
2. Reverse complement
DNA_ReverseComplement = {'A' : 'T', 'T': 'A', 'G':'C','C':'G'}
def reverse_complement(seq):
"""Swapping adenine with thymine and quanine with cytosine. Reversing newly generated string"""
return ''.join([DNA_ReverseComplement[nuc] for nuc in seq])复制
A clever way to use dictionary. Use the nucleotide we want to replace as the key, replacement as the value.
3. Comment your function
Already used it in the codes above, the triple quotes """{text}"""
are written under the def line, just to show what these functions are about.
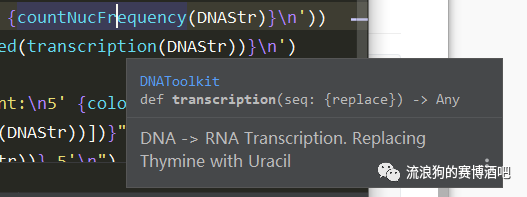
The amazing thing is, you can see the comments in other py files in which you import those functions when you put your mouse on the functions, just like it below:
4. Colorize the nucleotide output
def colored(seq):
bcolors = {
'A': '\033[92m',
'C': '\033[94m',
'G': '\033[93m',
'T': '\033[91m',
'U': '\033[91m',
'reset': '\033[0;0m'
}
tmpStr = ""
for nuc in seq:
if nuc in bcolors:
tmpStr += bcolors[nuc] + nuc
else:
tmpStr += bcolors['reset'] + nuc
return tmpStr + '\033[0;0m'复制
Not easy to use (mainly because of those complex color codes), but easy to read.
Use dictionary
and +=
to add color codes before nucleotides.
One thing to pay attention, after import this function, we also should rewrite our output with adding colored()
function.
5.Restructure and re-format the output
# DNA Toolset/Code testing file
from DNAToolkit import *
from utilities import colored
import random
# Creating a random DNA sequence for testing:
randDNAStr = ''.join([random.choice(Nucleotides)
for nuc in range(50)])
DNAStr = validateSeq(randDNAStr)
print(f'\nSequence:{colored(DNAStr)}\n')
print(f'[1] + Sequence Length: {len(DNAStr)}\n')
print(colored(f'[2] + Nucleotide Frequency: {countNucFrequency(DNAStr)}\n'))
print(f'[3] + DNA/RNA Transcription: {colored(transcription(DNAStr))}\n')
print(f"[4] + DNA String + Reverse Complement:\n5' {colored(DNAStr)} 3'")
print(f" {''.join(['|' for c in range(len(DNAStr))])}")
print(f"3' {colored(reverse_complement(DNAStr))} 5'\n")复制
We reformat the output to make the code more readable. If the file is too long with too many functions and steps, we may get lost. It's a good habit to add texts in every steps' output to explain what did we do in that step.
The f
letter is used for formatting strings. More straightforward, to add { } expression in strings.
This is the explain from python official docs.

One more thing to pay attention, we should add the colored
function before codes that print the string, otherwise the output won't be colored.
That's all for this course.
Reference
course video:https://www.youtube.com/watch?v=h1aP9HCFu6Y python docs: https://docs.python.org/3/tutorial/inputoutput.html python colorize archive: https://www.devdungeon.com/content/colorize-terminal-output-python
呜呜呜用英文写有点累以后还是用中文吧我自扇仨嘴巴子