16. PCA图绘制
清除当前环境中的变量
rm(list=ls())
设置工作目录
setwd("C:/Users/Dell/Desktop/R_Plots/16pca/")
加载示例数据
data <- read.table("demo_pca.txt",header = T,row.names = 1,sep="\t",check.names = F)
# 查看数据
head(data)
## C1 C2 C3 M1 M2 M3
## AT1G01060 33.035800 35.55930 34.64920 10.294500 13.393800 13.659000
## AT1G01320 73.740400 76.31340 69.04050 17.208400 19.669500 17.630300
## AT1G01420 6.667900 6.66516 7.94002 2.863710 3.834140 3.432600
## AT1G01520 9.588620 8.55635 9.32078 0.982357 2.876840 2.188840
## AT1G01970 17.210700 15.13220 18.35080 7.080410 7.748770 7.272220
## AT1G02380 0.749953 1.95807 1.44847 0.934699 0.367559 0.703017
# 数据转置,转换成行为样本,列为基因的矩阵
data <- t(data)
使用prcomp函数进行PCA分析
data.pca <- prcomp(data)
# 查看PCA的分析结果
summary(data.pca)
## Importance of components:
## PC1 PC2 PC3 PC4 PC5
## Standard deviation 1.751e+04 1.459e+03 426.25749 328.16697 171.7883
## Proportion of Variance 9.921e-01 6.890e-03 0.00059 0.00035 0.0001
## Cumulative Proportion 9.921e-01 9.990e-01 0.99956 0.99990 1.0000
## PC6
## Standard deviation 1.461e-11
## Proportion of Variance 0.000e+00
## Cumulative Proportion 1.000e+00
#----Standard deviation 标准差 其平方为方差=特征值
#----Proportion of Variance 方差贡献率
#----Cumulative Proportion 方差累计贡献率
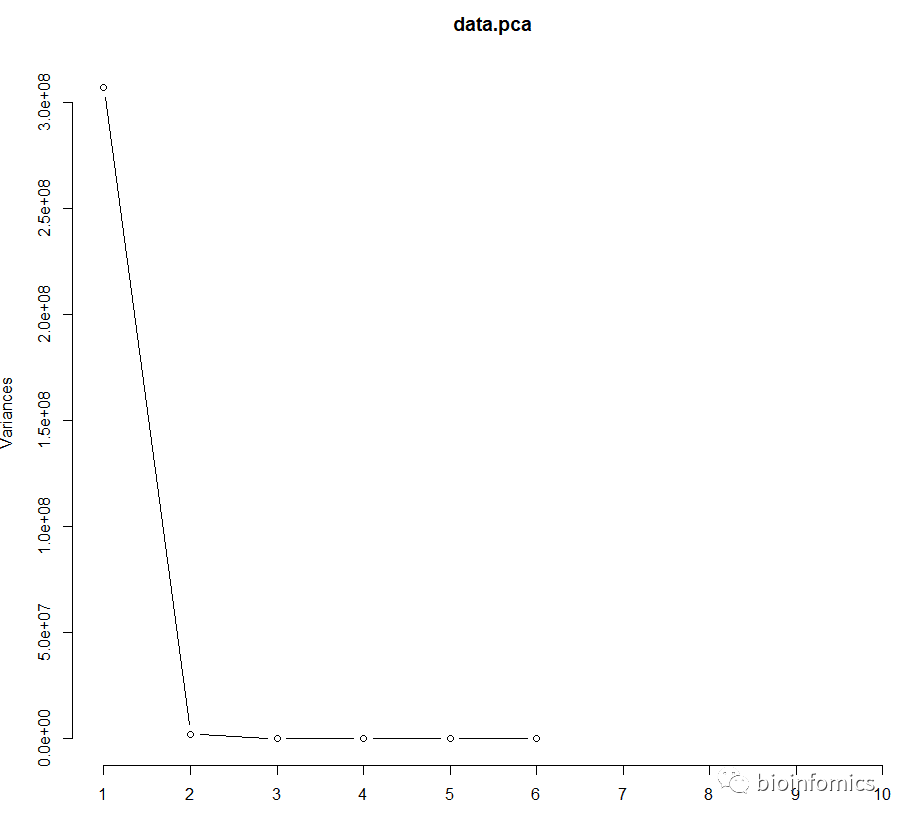
# 绘制主成分的碎石图
screeplot(data.pca, npcs = 10, type = "lines")
# 查看主成分的结果
head(data.pca$x)
## PC1 PC2 PC3 PC4 PC5 PC6
## C1 -13102.671 1379.8802 231.0448 148.3701 255.621964 -1.575094e-11
## C2 -15612.730 -1294.4214 -661.6586 137.3621 -1.050406 2.598617e-11
## C3 -15985.837 1072.3905 209.1377 -190.1187 -255.774746 1.695406e-11
## M1 16558.189 -442.8635 253.4135 517.0651 -100.838108 1.183409e-13
## M2 23877.097 1295.0204 -408.9296 -249.7849 24.401970 -4.182591e-11
## M3 4265.951 -2010.0062 376.9922 -362.8937 77.639326 1.456456e-11
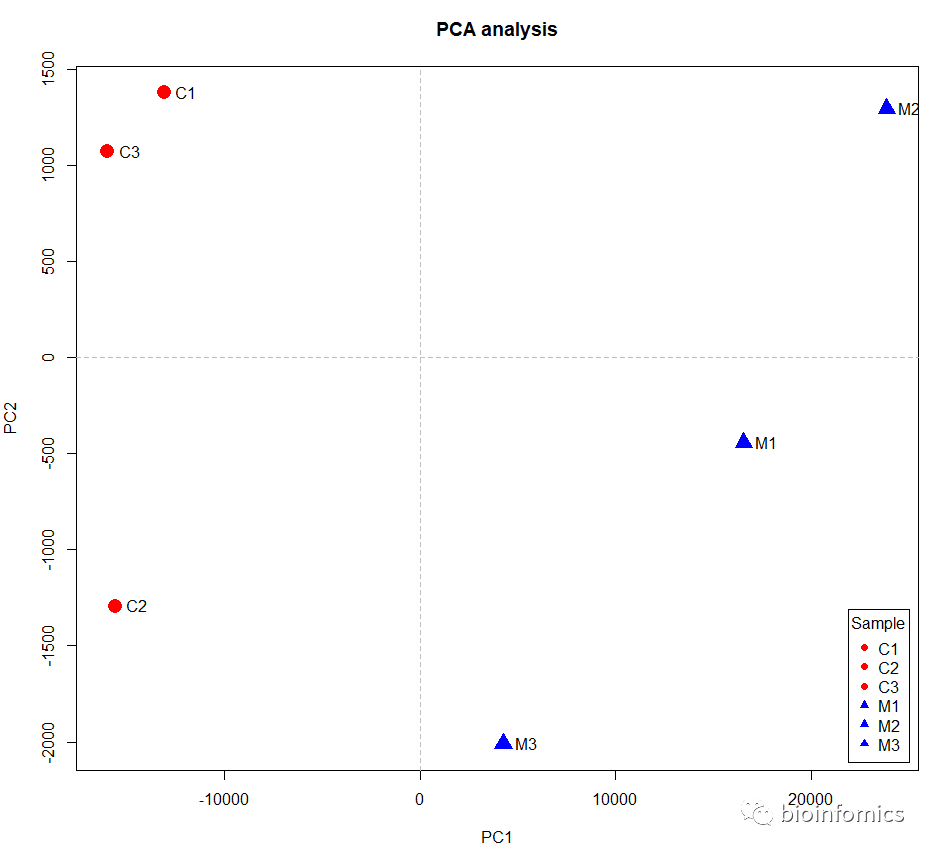
使用基础plot函数绘制PCA图
plot(data.pca$x,cex = 2,main = "PCA analysis",
col = c(rep("red",3),rep("blue",3)),
pch = c(rep(16,3),rep(17,3)))
# 添加分隔线
abline(h=0,v=0,lty=2,col="gray")
# 添加标签
text(data.pca$x,labels = rownames(data.pca$x),pos = 4,offset = 0.6,cex = 1)
# 添加图例
legend("bottomright",title = "Sample",inset = 0.01,
legend = rownames(data.pca$x),
col = c(rep("red",3),rep("blue",3)),
pch = c(rep(16,3),rep(17,3)))
使用ggplot2包绘制PCA图
library(ggplot2)
# 查看示例数据
head(USArrests)
## Murder Assault UrbanPop Rape
## Alabama 13.2 236 58 21.2
## Alaska 10.0 263 48 44.5
## Arizona 8.1 294 80 31.0
## Arkansas 8.8 190 50 19.5
## California 9.0 276 91 40.6
## Colorado 7.9 204 78 38.7
# 使用princomp函数进行PCA分析
data.pca <- princomp(USArrests,cor = T)
# 查看PCA的结果
summary(data.pca)
## Importance of components:
## Comp.1 Comp.2 Comp.3 Comp.4
## Standard deviation 1.5748783 0.9948694 0.5971291 0.41644938
## Proportion of Variance 0.6200604 0.2474413 0.0891408 0.04335752
## Cumulative Proportion 0.6200604 0.8675017 0.9566425 1.00000000
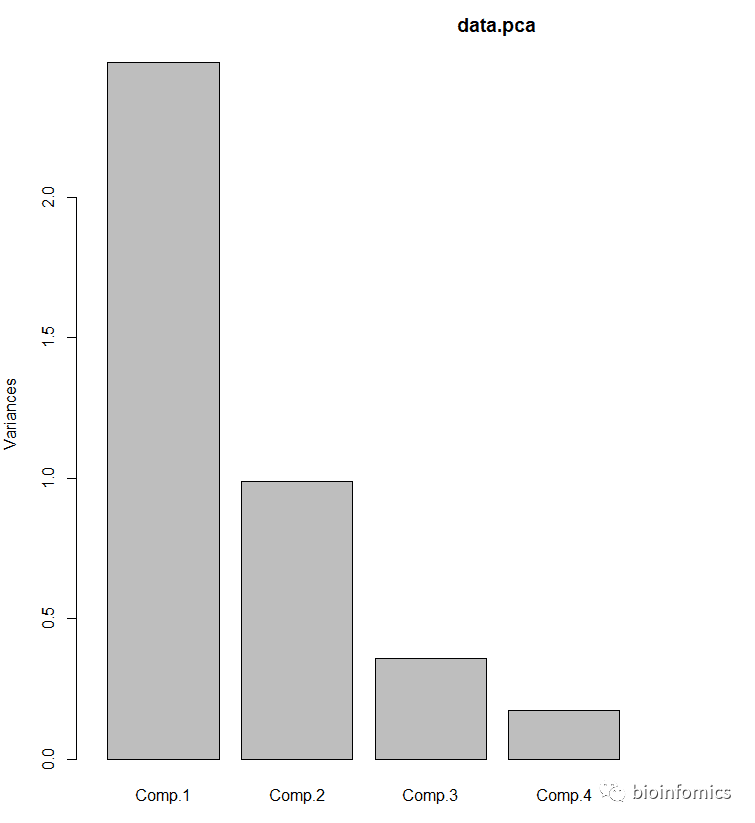
# 绘制主成分碎石图
screeplot(data.pca,npcs = 6,type = "barplot")
#查看主成分的结果
pca.scores <- as.data.frame(data.pca$scores)
head(pca.scores)
## Comp.1 Comp.2 Comp.3 Comp.4
## Alabama 0.9855659 1.1333924 0.44426879 0.156267145
## Alaska 1.9501378 1.0732133 -2.04000333 -0.438583440
## Arizona 1.7631635 -0.7459568 -0.05478082 -0.834652924
## Arkansas -0.1414203 1.1197968 -0.11457369 -0.182810896
## California 2.5239801 -1.5429340 -0.59855680 -0.341996478
## Colorado 1.5145629 -0.9875551 -1.09500699 0.001464887
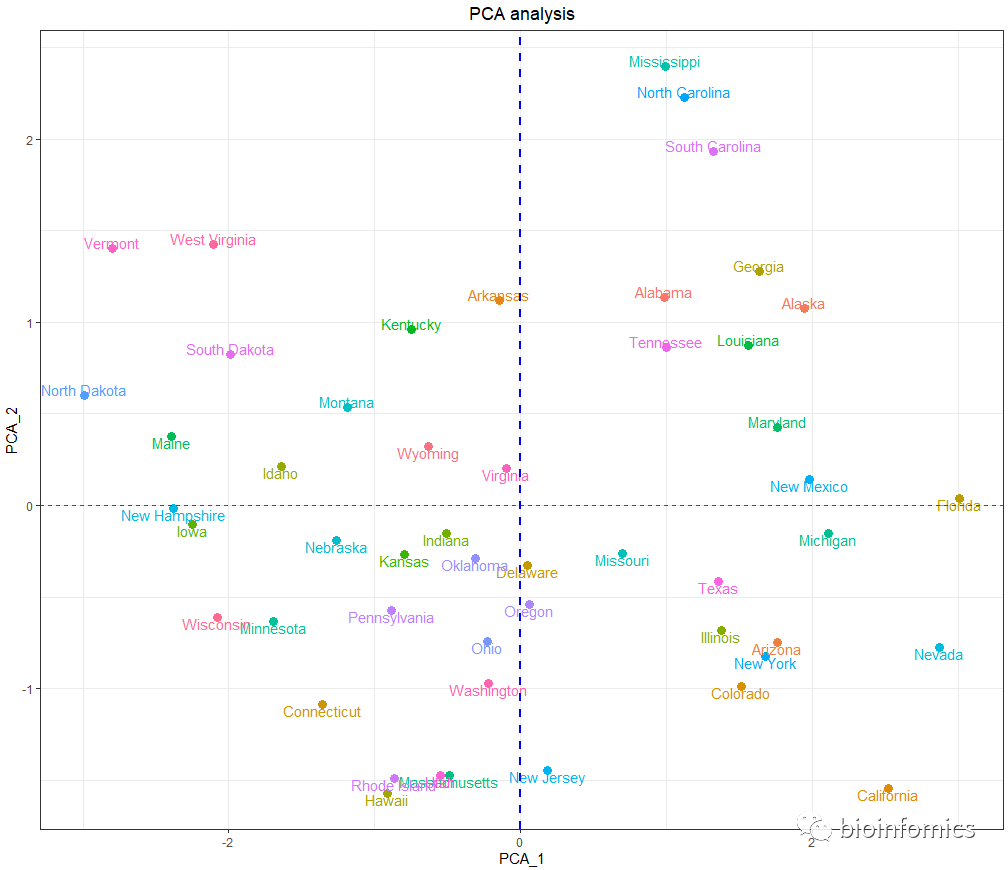
# 绘制PCA图
ggplot(pca.scores,aes(Comp.1,Comp.2,col=rownames(pca.scores))) +
geom_point(size=3) +
geom_text(aes(label=rownames(pca.scores)),vjust = "outward") +
geom_hline(yintercept = 0,lty=2,col="red") +
geom_vline(xintercept = 0,lty=2,col="blue",lwd=1) +
theme_bw() + theme(legend.position = "none") +
theme(plot.title = element_text(hjust = 0.5)) +
labs(x="PCA_1",y="PCA_2",title = "PCA analysis")
使用scatterplot3d包绘制三维PCA图
library(scatterplot3d)
# 加载示例数据
data <- read.table("demo_pca.txt",header = T,row.names = 1,sep="\t",check.names = F)
head(data)
## C1 C2 C3 M1 M2 M3
## AT1G01060 33.035800 35.55930 34.64920 10.294500 13.393800 13.659000
## AT1G01320 73.740400 76.31340 69.04050 17.208400 19.669500 17.630300
## AT1G01420 6.667900 6.66516 7.94002 2.863710 3.834140 3.432600
## AT1G01520 9.588620 8.55635 9.32078 0.982357 2.876840 2.188840
## AT1G01970 17.210700 15.13220 18.35080 7.080410 7.748770 7.272220
## AT1G02380 0.749953 1.95807 1.44847 0.934699 0.367559 0.703017
# 数据转置,转换成行为样本,列为基因的矩阵
data <- t(data)
# 使用prcomp函数进行PCA分析
data.pca <- prcomp(data)
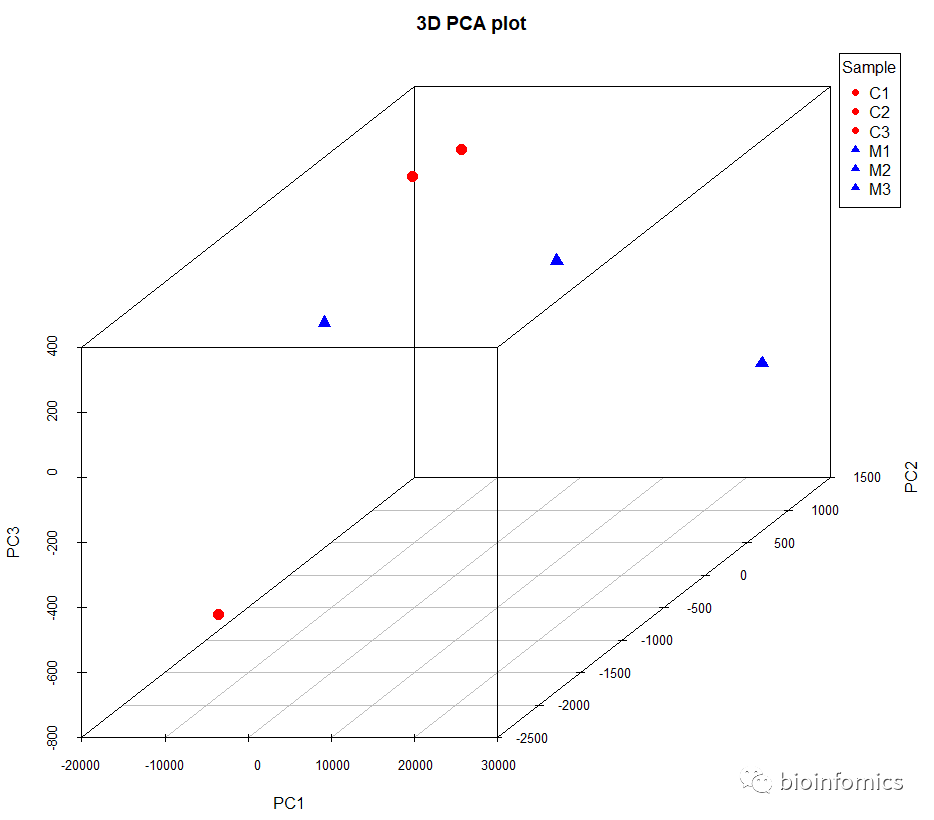
# 绘制三维PCA图
scatterplot3d(data.pca$x[,1:3],
pch = c(rep(16,3),rep(17,3)),
color= c(rep("red",3),rep("blue",3)),
angle=45, main= "3D PCA plot",
cex.symbols= 1.5,,mar=c(5, 4, 4, 5))
# 添加图例
legend("topright",title = "Sample",
xpd=TRUE,inset= -0.01,
legend = rownames(data.pca$x),
col = c(rep("red",3),rep("blue",3)),
pch = c(rep(16,3),rep(17,3)))
使用factoextra包绘制PCA图
library(factoextra)
# 查看示例数据
head(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
# 使用prcomp函数进行PCA分析
res.pca <- prcomp(iris[, -5], scale = TRUE)
res.pca
## Standard deviations (1, .., p=4):
## [1] 1.7083611 0.9560494 0.3830886 0.1439265
##
## Rotation (n x k) = (4 x 4):
## PC1 PC2 PC3 PC4
## Sepal.Length 0.5210659 -0.37741762 0.7195664 0.2612863
## Sepal.Width -0.2693474 -0.92329566 -0.2443818 -0.1235096
## Petal.Length 0.5804131 -0.02449161 -0.1421264 -0.8014492
## Petal.Width 0.5648565 -0.06694199 -0.6342727 0.5235971
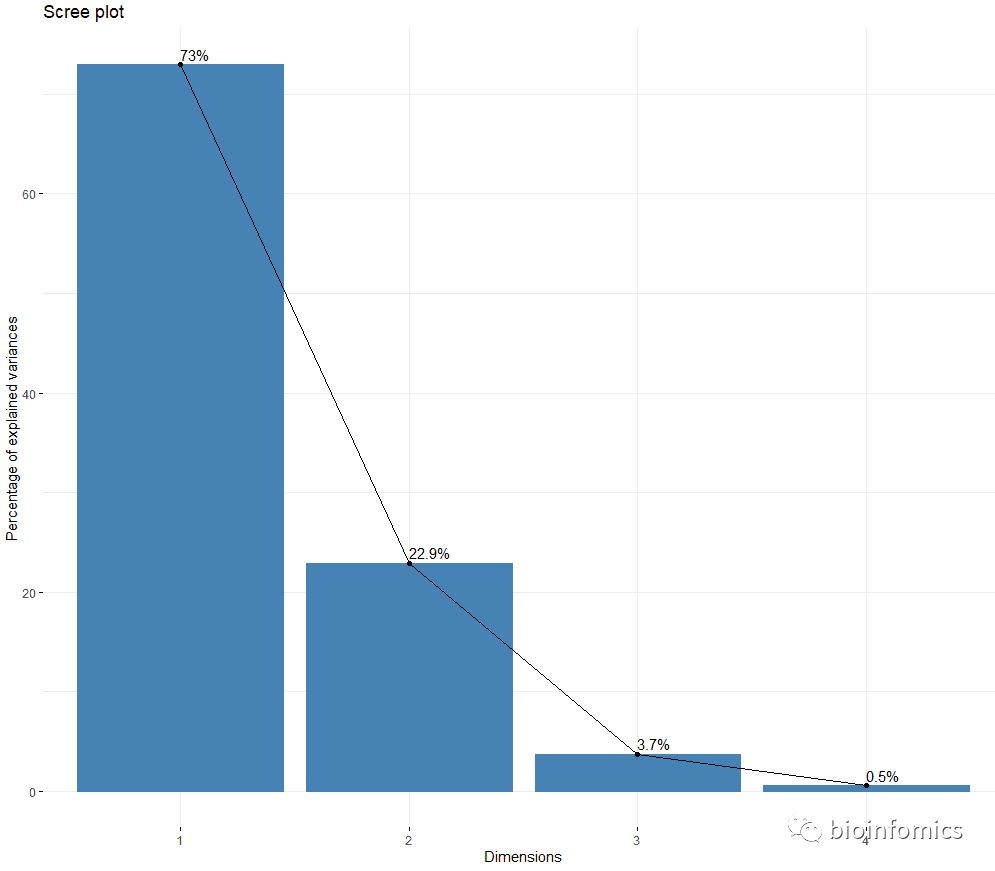
#绘制主成分碎石图
fviz_screeplot(res.pca, addlabels = TRUE)
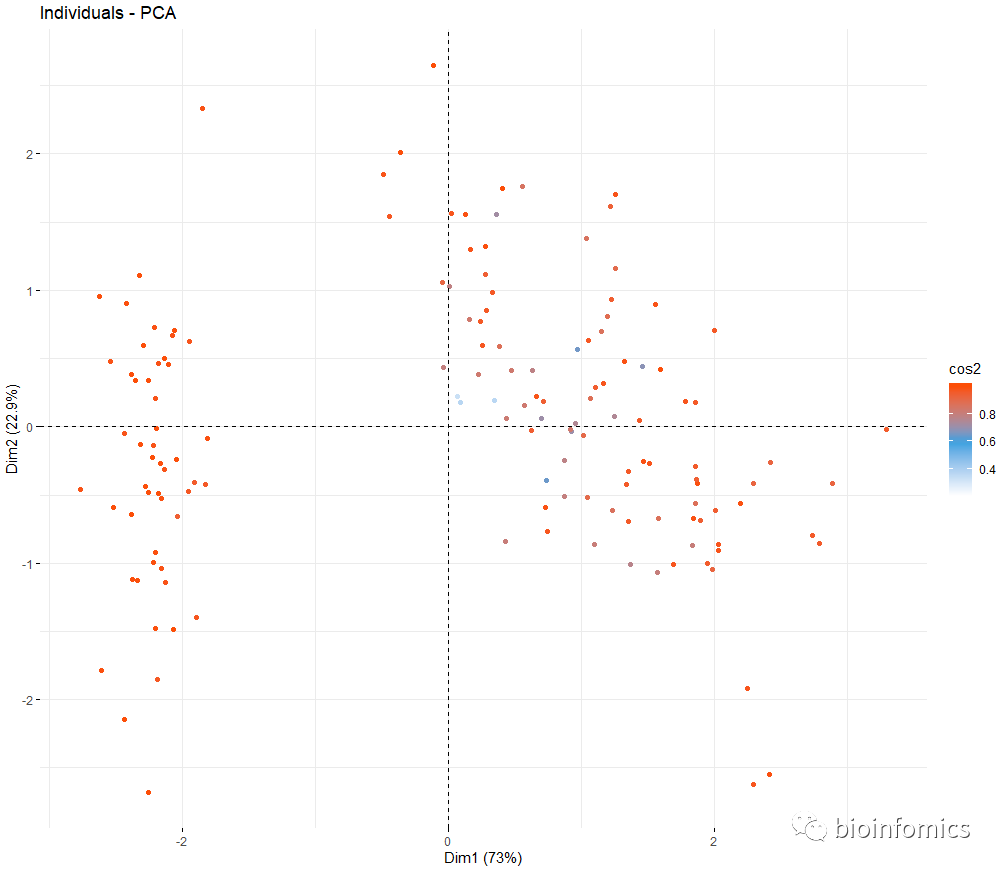
# 可视化PCA分析的结果
# Graph of individuals
# +++++++++++++++++++++
fviz_pca_ind(res.pca, col.ind="cos2",
geom = "point", # show points only
gradient.cols = c("white", "#2E9FDF", "#FC4E07" ))
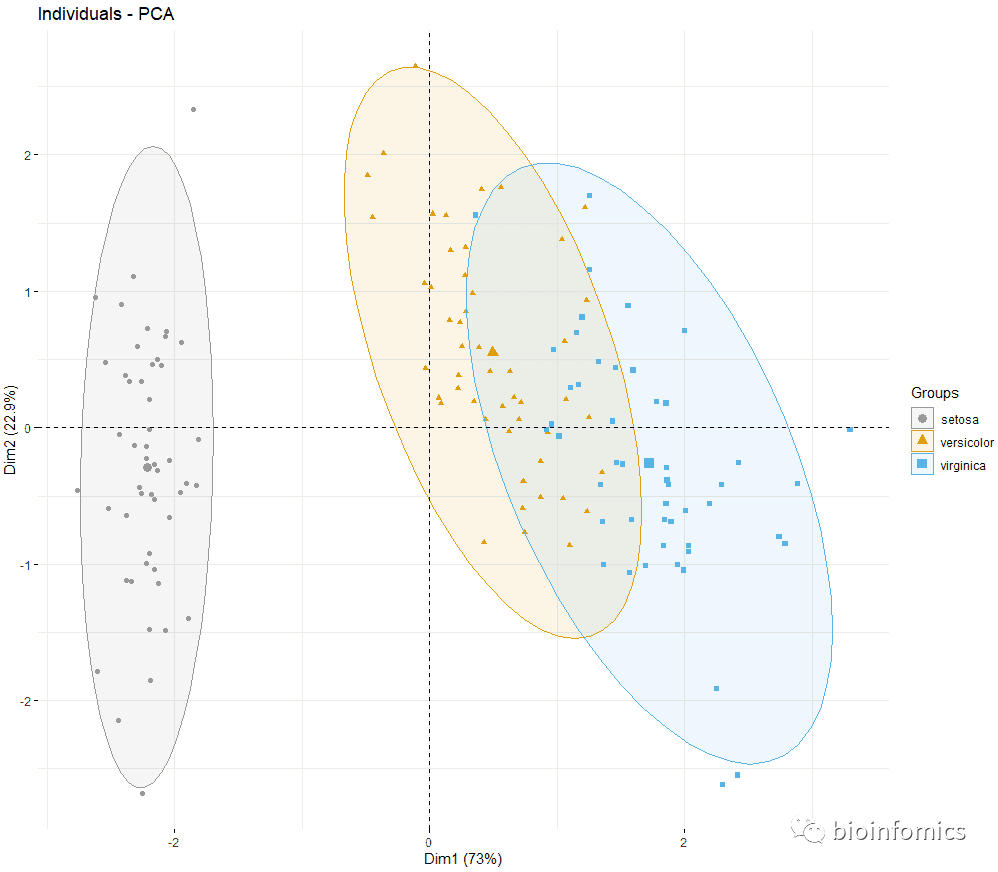
fviz_pca_ind(res.pca, label="none", habillage=iris$Species,
addEllipses=TRUE, ellipse.level=0.95,
palette = c("#999999", "#E69F00", "#56B4E9"))
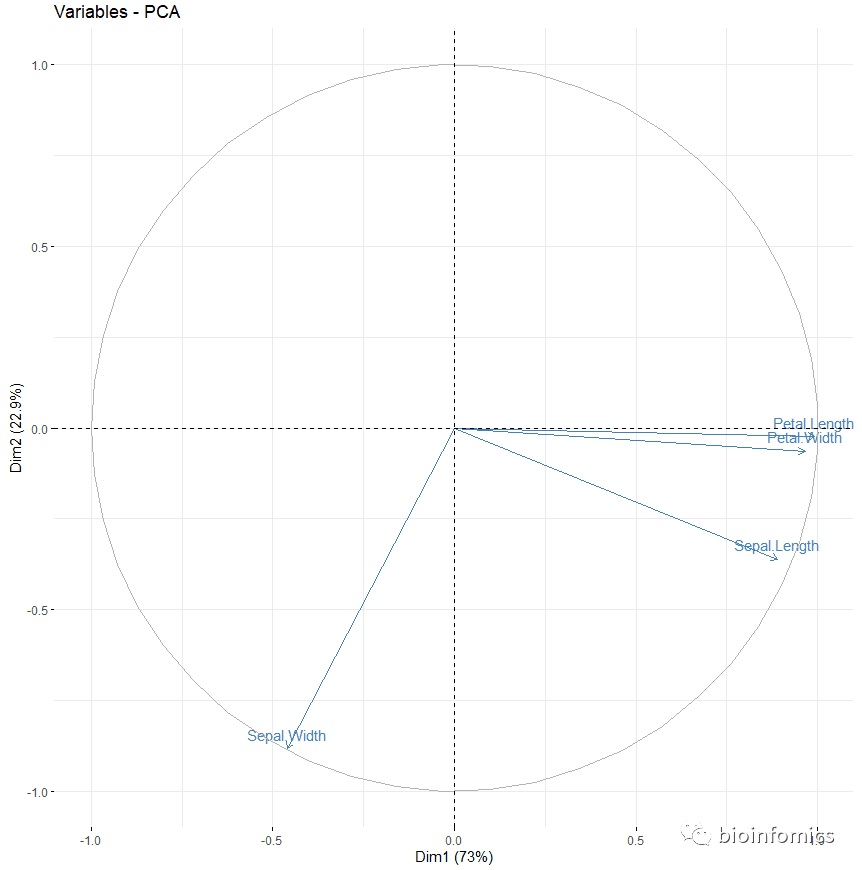
# Graph of variables
# +++++++++++++++++++++
# Default plot
fviz_pca_var(res.pca, col.var = "steelblue")
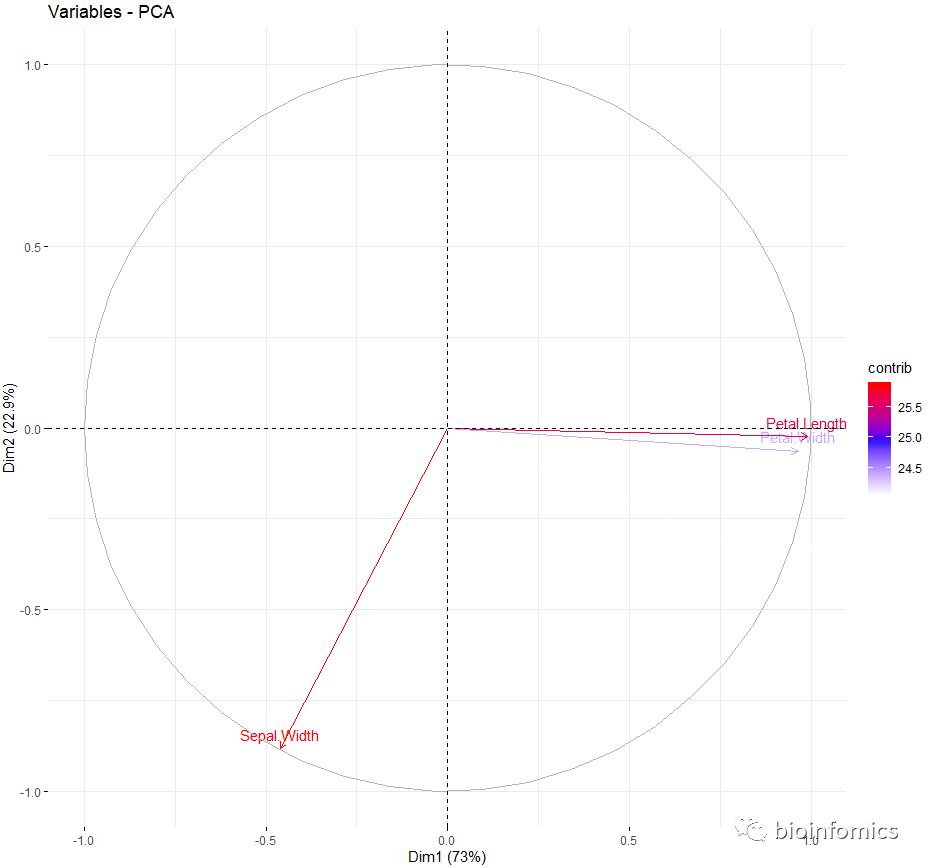
# Control variable colors using their contributions
fviz_pca_var(res.pca, col.var = "contrib",
gradient.cols = c("white", "blue", "red"),
ggtheme = theme_minimal())
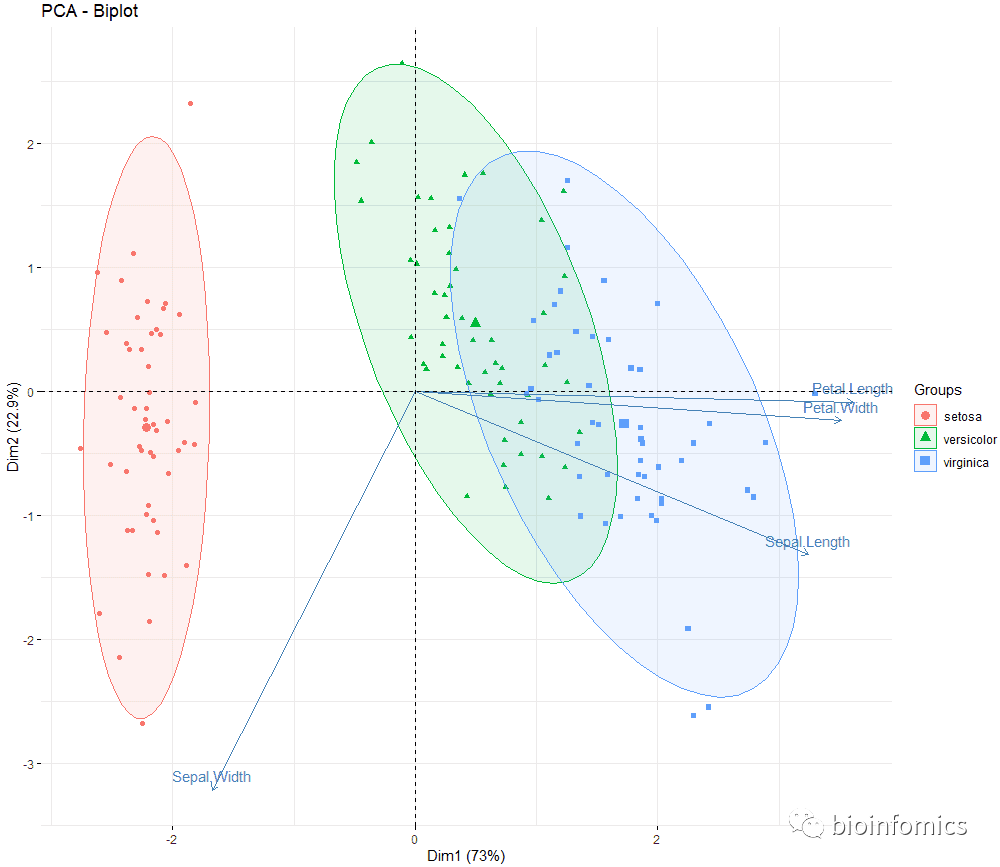
# Biplot of individuals and variables
# +++++++++++++++++++++
# Keep only the labels for variables
# Change the color by groups, add ellipses
fviz_pca_biplot(res.pca, label = "var", habillage=iris$Species,
addEllipses=TRUE, ellipse.level=0.95,
ggtheme = theme_minimal())
sessionInfo()
R version 3.6.0 (2019-04-26)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18363)
Matrix products: default
locale:
[1] LC_COLLATE=Chinese (Simplified)_China.936
[2] LC_CTYPE=Chinese (Simplified)_China.936
[3] LC_MONETARY=Chinese (Simplified)_China.936
[4] LC_NUMERIC=C
[5] LC_TIME=Chinese (Simplified)_China.936
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] FactoMineR_2.3 factoextra_1.0.7 scatterplot3d_0.3-41
[4] ggplot2_3.2.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 pillar_1.4.2 compiler_3.6.0 RColorBrewer_1.1-2
[5] ggpubr_0.2.1 tools_3.6.0 digest_0.6.20 evaluate_0.14
[9] tibble_2.1.3 gtable_0.3.0 lattice_0.20-38 pkgconfig_2.0.2
[13] rlang_0.4.7 rstudioapi_0.10 ggrepel_0.8.1 yaml_2.2.0
[17] xfun_0.8 knitr_1.23 withr_2.1.2 dplyr_0.8.3
[21] cluster_2.0.8 flashClust_1.01-2 grid_3.6.0 tidyselect_0.2.5
[25] glue_1.3.1 R6_2.4.0 rmarkdown_1.13 purrr_0.3.2
[29] magrittr_1.5 htmltools_0.3.6 scales_1.0.0 leaps_3.1
[33] MASS_7.3-51.4 rsconnect_0.8.16 assertthat_0.2.1 colorspace_1.4-1
[37] ggsignif_0.5.0 labeling_0.3 lazyeval_0.2.2 munsell_0.5.0
[41] crayon_1.3.4

END






文章转载自bioinfomics,如果涉嫌侵权,请发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。