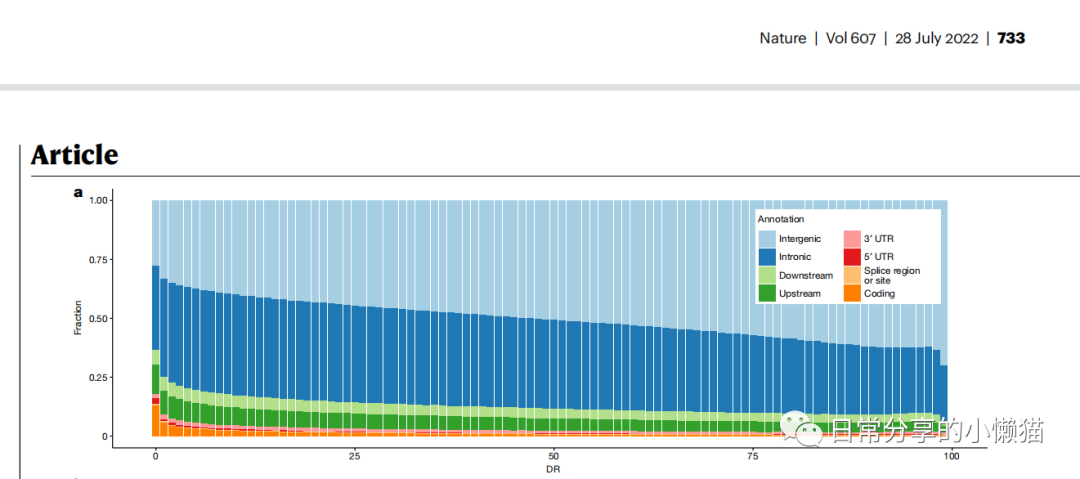
本文主要展示如何绘制来自nature期刊The sequences of 150,119 genomes in the UK Biobank(Halldorsson, B.V.,et al,2022) [1] 一文中的Fig.2a条形图。该条形图用于展示基因系列的变化状况,如下图所示:
1、数据准备
library(dplyr)
library(ggplot2)
# 构造数据
mydata <-
data.frame(
DR = 1:100,
value.A = seq(1201,2000,8),
value.B = seq(1200,1101,-1),
value.C = 600:501,
value.D = 500:401,
value.E = 400:301,
value.F = 300:201,
value.G = 200:101,
value.H = 100:1) %>%
reshape2::melt(id.vars = "DR", value.name = "value", variable.name = "tpye")
#查看数据
head(mydata)
# DR tpye value
#1 1 value.A 1201
#2 2 value.A 1209
#3 3 value.A 1217
#4 4 value.A 1225
#5 5 value.A 1233
#6 6 value.A 1241
str(mydata)
#'data.frame': 800 obs. of 3 variables:
# $ DR : int 1 2 3 4 5 6 7 8 9 10 ...
# $ tpye : Factor w/ 8 levels "value.A","value.B",..: 1 1 1 1 1 1 1 1 1 1 ...
# $ value: num 1201 1209 1217 1225 1233 ...
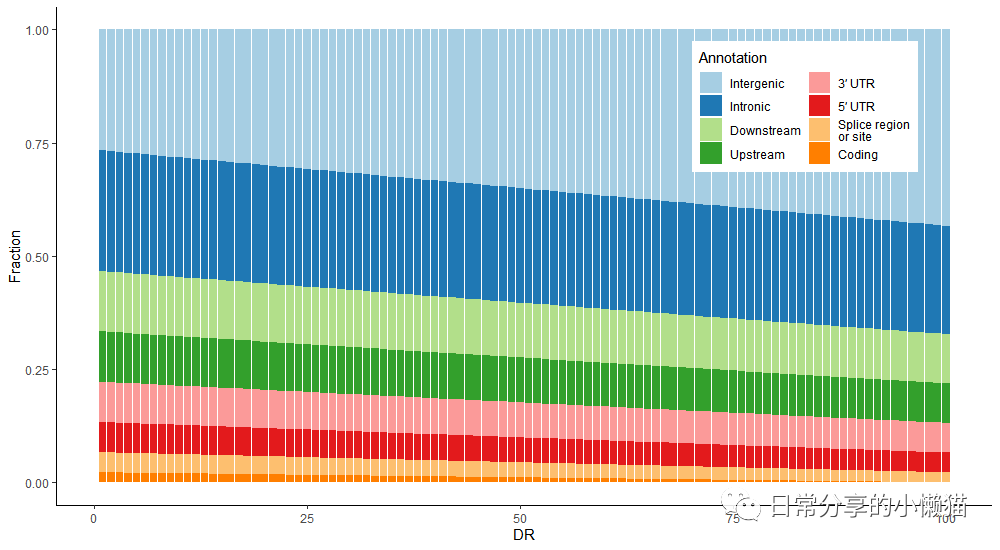
2、图形绘制
#生成颜色
mycolor <- c("#A6CEE3", "#1F78B4", "#B2DF8A", "#33A02C", "#FB9A99", "#E31A1C", "#FDBF6F", "#FF7F00")
#绘图
ggplot(mydata,aes(DR, value, fill = tpye)) +
geom_col(position = "fill") +
scale_fill_manual(values = mycolor, labels = c("Intergenic", "Intronic", "Downstream", "Upstream",
"3′ UTR", "5′ UTR", "Splice region\nor site", "Coding")) +
theme_classic() +
labs(x = "DR" , y = "Fraction", fill = "Annotation") +
guides(fill = guide_legend(ncol = 2)) +
theme(legend.position = c(0.8, 0.8))
3、其他
其他绘图方法可进一步阅读公众号其他文章。
如有帮助请多多点赞哦!
参考资料
[1] Halldorsson, B.V., Eggertsson, H.P., Moore, K.H.S. et al. The sequences of 150,119 genomes in the UK Biobank. Nature 607, 732–740 (2022). https://doi.org/10.1038/s41586-022-04965-x
文章转载自日常分享的小懒猫,如果涉嫌侵权,请发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。