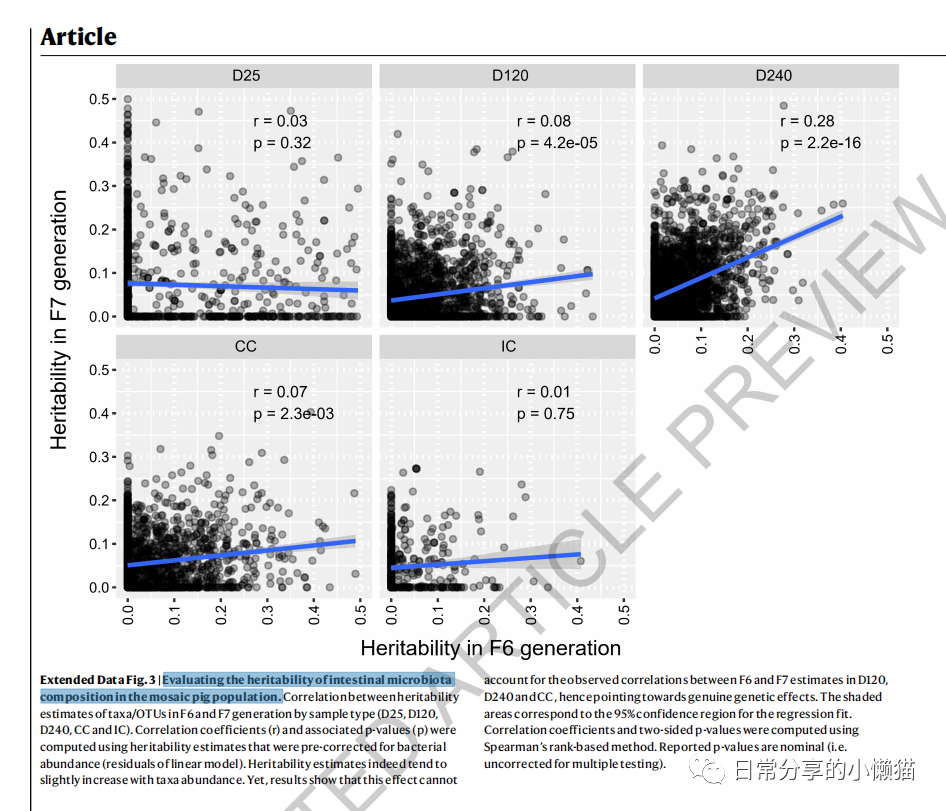
本文主要展示如何利用R语言绘制来自nature期刊ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs(Hui Yang, et al,2022)[1]一文中的Extended Data Fig.3。该图形式为散点图,并添加了拟合曲线以及相关性系数r及对应p值,用于展示样本之间的关联性。如下图所示:
1、图形绘制
library(ggpubr)
library(ggplot2)
library(ggpmisc)
library(dplyr)
her_out <- read.delim("result.txt")
her_out <- her_out %>% dplyr::select(Sample_type,F6_her ,F7_her)
her_out$Sample_type <- factor(her_out$Sample_type, levels = c("D25","D120","D240","CC","IC"))
head(her_out)
# Sample_type F6_her F7_her
#1 D25 0.0000 NA
#2 D120 0.0000 NA
#3 D240 0.0315 NA
#4 IC 0.0000 NA
#5 D25 0.0000 NA
#6 D240 0.0297 NA
str(her_out)
#'data.frame': 17223 obs. of 3 variables:
# $ Sample_type: Factor w/ 5 levels "D25","D120","D240",..: 1 2 3 5 1 3 5 5 1 3 ...
# $ F6_her : num 0 0 0.0315 0 0 0.0297 0 0 0 0.0215 ...
# $ F7_her : num NA NA NA NA NA NA NA NA NA NA ...
# 图形绘制
ggplot(her_out, aes(F6_her, F7_her)) +
geom_point(alpha = 0.5) +
stat_cor(p.accuracy = 0.01, r.accuracy = 0.01, method = "pearson") + #spearman
geom_smooth(method = "lm") +
scale_y_continuous(breaks = seq(0, 0.6, 0.1)) +
scale_x_continuous(breaks = seq(0, 0.6, 0.1)) +
facet_wrap(~Sample_type) +
theme_bw() +
theme(axis.text = element_text(size = 12),
strip.text = element_text(size = 12),
axis.text.x = element_text(angle = 90, vjust = 0.5),
axis.title = element_text(size = 14),
panel.background = element_rect(fill = "#f0f0f0"),
panel.grid.major = element_line(color = "white", linetype = "dotted", size = 1)) +
labs(x = "Heritability in F6 generation", y = "Heritability in F7 generation")
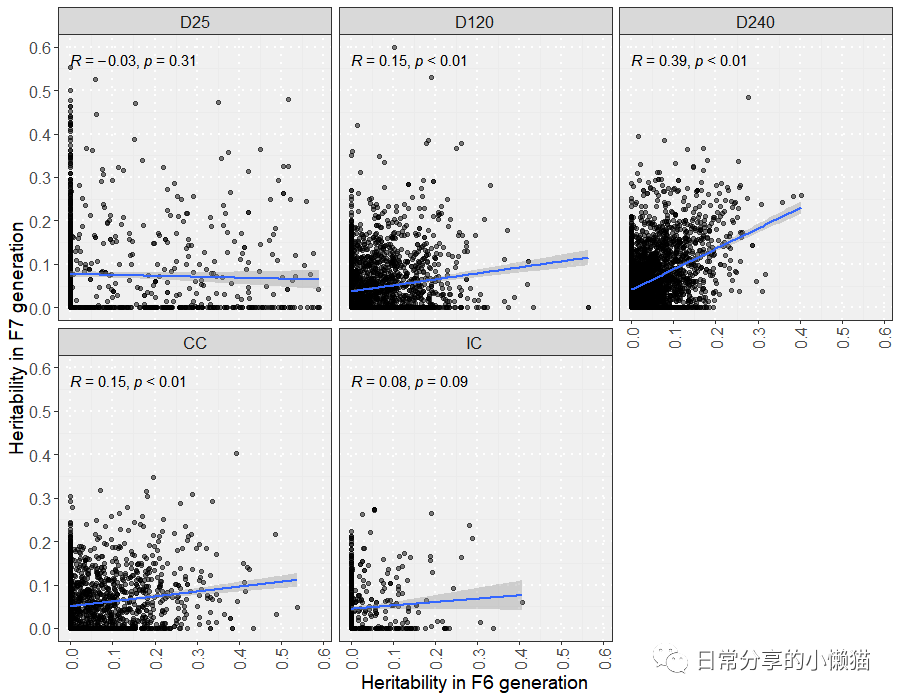
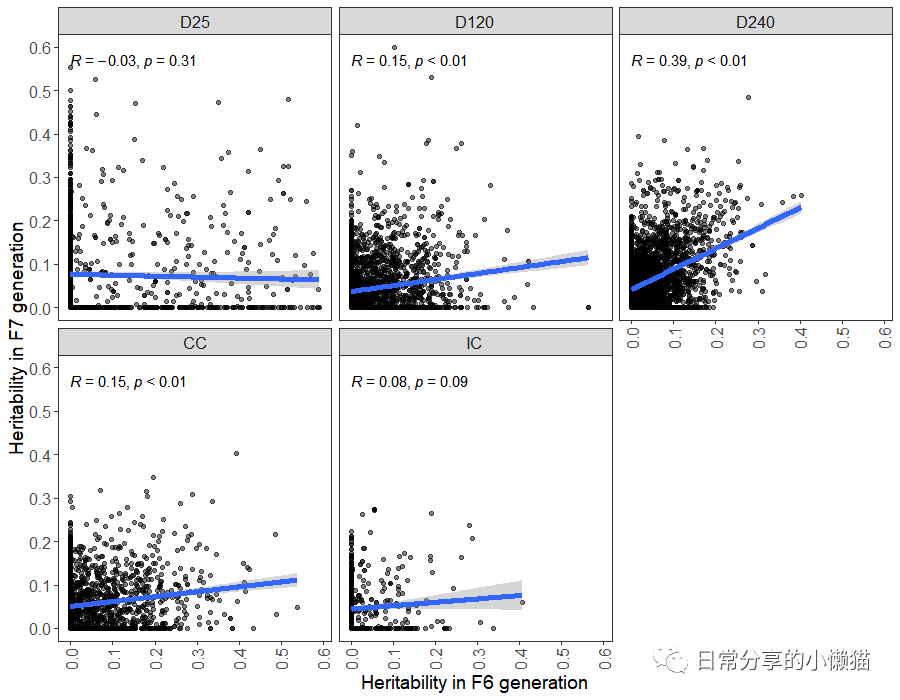
该图主要使用geom_point() 函数绘制散点图,使用geom_smooth() 函数添加拟合曲线,使用ggpubr包中的 stat_cor() 函数来添加相关系数及p值,并利用facet_wrap() 函数进行分面。
2、进一步调整
修改主题
ggplot(her_out, aes(F6_her, F7_her)) +
geom_point(alpha = 0.5) +
stat_cor(p.accuracy = 0.01, r.accuracy = 0.01, method = "pearson") + #spearman
geom_smooth(method = "lm", size = 2) +
scale_y_continuous(breaks = seq(0, 0.6, 0.1)) +
scale_x_continuous(breaks = seq(0, 0.6, 0.1)) +
facet_wrap(~Sample_type) +
theme_bw() +
theme(axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 90, vjust = 0.5),
strip.text = element_text(size = 12),
axis.title = element_text(size = 14),
panel.grid = element_blank()) +
labs(x = "Heritability in F6 generation", y = "Heritability in F7 generation")
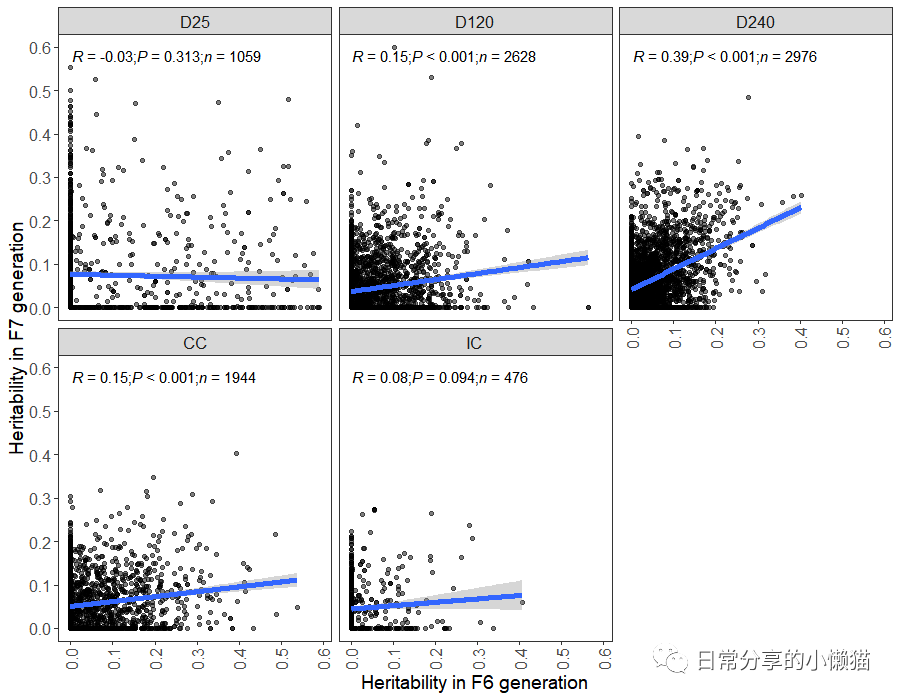
使用ggpmisc包添加相关系数
此外,除了使用ggpubr包来添加相关系数外,还可以使用ggpmisc包中的stat_correlation() 函数来添加变量相关性及p值。
ggplot(her_out, aes(F6_her, F7_her)) +
geom_point(alpha = 0.5) +
stat_correlation(aes(label = paste(after_stat(cor.label), after_stat(p.value.label), after_stat(n.label), sep = '*";"*')),
method = "pearson") +
geom_smooth(method = "lm", size = 2) +
scale_y_continuous(breaks = seq(0, 0.6, 0.1)) +
scale_x_continuous(breaks = seq(0, 0.6, 0.1)) +
facet_wrap(~Sample_type) +
theme_bw() +
theme(axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 90, vjust = 0.5),
strip.text = element_text(size = 12),
axis.title = element_text(size = 14),
panel.grid = element_blank()) +
labs(x = "Heritability in F6 generation", y = "Heritability in F7 generation")
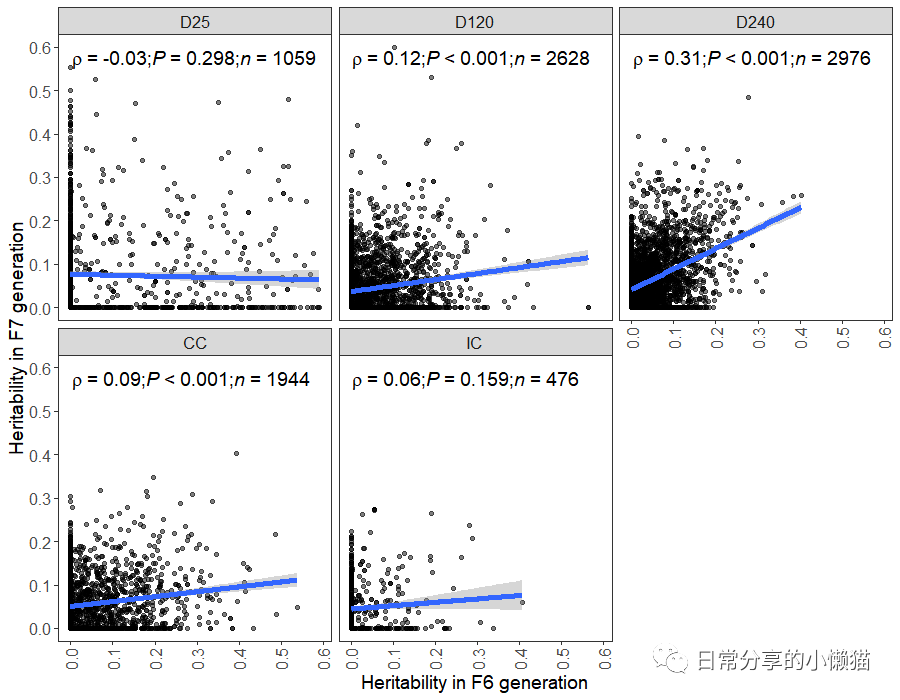
使用spearman相关系数
ggplot(her_out, aes(F6_her, F7_her)) +
geom_point(alpha = 0.5) +
stat_correlation(aes(label = paste(after_stat(r.label), after_stat(p.value.label), after_stat(n.label), sep = '*";"*')),
method = "spearman", size = 5) + # spearman、pearson
geom_smooth(method = "lm", size = 2) +
scale_y_continuous(breaks = seq(0, 0.6, 0.1)) +
scale_x_continuous(breaks = seq(0, 0.6, 0.1)) +
facet_wrap(~Sample_type) +
theme_bw() +
theme(axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 90, vjust = 0.5),
strip.text = element_text(size = 12),
axis.title = element_text(size = 14),
panel.grid = element_blank()) +
labs(x = "Heritability in F6 generation", y = "Heritability in F7 generation")
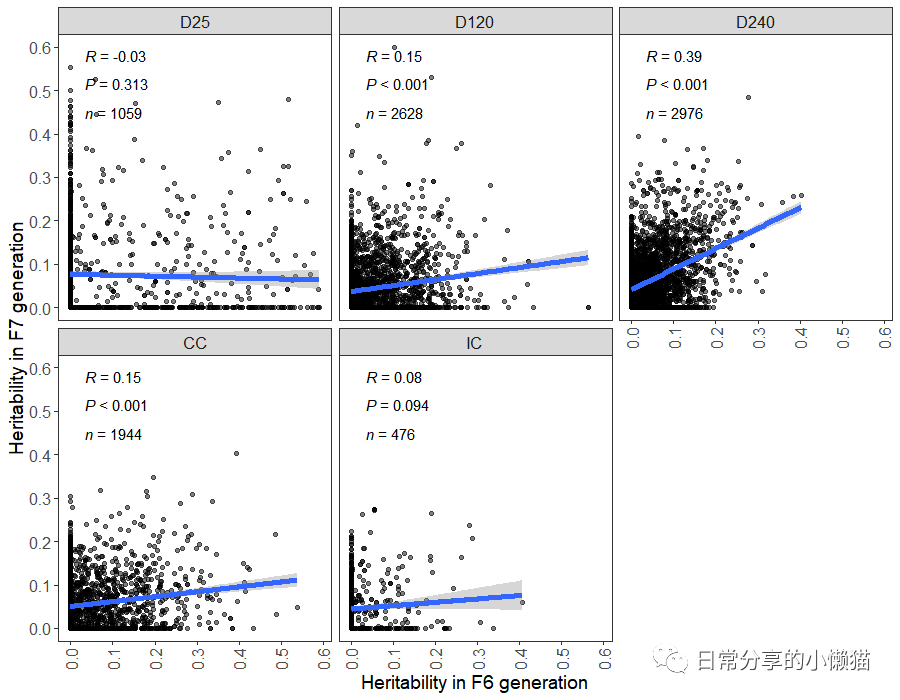
将显著性及p值拆分
ggplot(her_out, aes(F6_her, F7_her)) +
geom_point(alpha = 0.5) +
stat_correlation(aes(label = after_stat(cor.label), method = "pearson"), label.x = 0.1, label.y = 0.95) + # spearman、pearson
stat_correlation(aes(label = after_stat(p.value.label), method = "pearson"), label.x = 0.1, label.y = 0.85) +
stat_correlation(aes(label = after_stat(n.label)), label.x = 0.1, label.y = 0.75) +
geom_smooth(method = "lm", size = 2) +
scale_y_continuous(breaks = seq(0, 0.6, 0.1)) +
scale_x_continuous(breaks = seq(0, 0.6, 0.1)) +
facet_wrap(~Sample_type) +
theme_bw() +
theme(axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 90, vjust = 0.5),
strip.text = element_text(size = 12),
axis.title = element_text(size = 14),
panel.grid = element_blank()) +
labs(x = "Heritability in F6 generation", y = "Heritability in F7 generation")
3、其他
其他绘图方法可进一步阅读公众号其他文章。
如有帮助请多多点赞哦!
参考资料
Yang, H., Wu, J., Huang, X. et al. ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature 606, 358–367 (2022). : https://doi.org/10.1038/s41586-022-04769-z
文章转载自日常分享的小懒猫,如果涉嫌侵权,请发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。