




ICDE2023_Decision Support System for Chronic Diseases Based on Drug-Drug Interactions_腾讯云.pdf
免费下载
Decision Support System for Chronic Diseases
Based on Drug-Drug Interactions
Tian Bian
1
, Yuli Jiang
1
, Jia Li
2*
, Tingyang Xu
3*
, Yu Rong
3
,
Yi Su
4
, Timothy Kwok
1
, Helen Meng
1
, Hong Cheng
1
1
The Chinese University of Hong Kong,
2
Hong Kong University of Science and Technology (Guangzhou),
3
Tencent AI Lab,
4
Hunan Normal University
{tbian,hmmeng,hcheng}@se.cuhk.edu.hk, yljiang@cse.cuhk.edu.hk, jialee@ust.hk,
tingyangxu@tencent.com, yu.rong@hotmail.com, alddle@hunnu.edu.cn, tkwok@cuhk.edu.hk
Abstract—Many patients with chronic diseases resort to mul-
tiple medications to relieve various symptoms, which raises
concerns about the safety of multiple medication use, as severe
drug-drug antagonism can lead to serious adverse effects or even
death. This paper presents a Decision Support System, called
DSSDDI, based on drug-drug interactions to support doctors
prescribing decisions. DSSDDI contains three modules, Drug-
Drug Interaction (DDI) module, Medical Decision (MD) module
and Medical Support (MS) module. The DDI module learns
safer and more effective drug representations from the drug-
drug interactions. To capture the potential causal relationship
between DDI and medication use, the MD module considers
the representations of patients and drugs as context, DDI and
patients’ similarity as treatment, and medication use as outcome
to construct counterfactual links for the representation learning.
Furthermore, the MS module provides drug candidates to doctors
with explanations. Experiments on the chronic data collected
from the Hong Kong Chronic Disease Study Project and a public
diagnostic data MIMIC-III demonstrate that DSSDDI can be a
reliable reference for doctors in terms of safety and efficiency
of clinical diagnosis, with significant improvements compared
to baseline methods. Source code of the proposed DSSDDI is
publicly available at https://github.com/TianBian95/DSSDDI.
Index Terms—Decision Support System, Drug-Drug Interac-
tions, Causal Inference
I. INTRODUCTION
Due to physiological changes, increased risk of disease
and decreased drug clearance, problematic polypharmacy has
become a significant factor in the increased risk of severe Ad-
verse Drug Events (ADEs), hospital admissions, and death in
chronic patients [1]. This issue is especially prominent during
critical times, such as the epidemic of coronavirus disease
(COVID-19). With the shortage of clinical resources, med-
ication for chronic patients lacks guidance from professional
doctors and presents unique challenges. Further, polypharmacy
increases the potential for drug-drug interactions (DDI), in-
cluding potentially inappropriate drug combinations present in
prescription medications [2], which accelerates the imbalance
between the complex needs of the chronic patients and the
problems caused by the multiple medications. A systematic
approach is required to efficiently support doctors in tailoring
∗
Corresponding authors.
of medication regimens to extricate the chronic patients from
the dilemma.
Advanced technologies nowadays have been applied to
develop more effective decision support systems for better-
informed decisions [3]. However, some methods [4], [5] that
learn from association between patients and drugs, mainly
rely on patient and drug features, but ignore the impact of
DDI on medical decisions. Some other methods [6] make
use of DDI to learn drug embeddings but fail to capture the
causal relationship between drug embeddings and medication
suggestions. Therefore, our goal in this paper is: (1) leverag-
ing DDI to avoid severe ADEs in medication suggestions,
and (2) employing the causal model [7], [8] to learn the
potential causal relationships between DDI and medication
suggestions to improve the accuracy.
We studied the chronic patients through the Hong Kong
Chronic Disease Study Project, including their personal fea-
tures, clinical history, psychological assessment and medica-
tion use. We propose a decision support system, DSSDDI, that
can provide explainable medication suggestions for chronic
diseases. A bipartite graph can be naturally formed on the
patients and drugs, and then DSSDDI applies link prediction
on the bipartite graph for medication suggestions. With the
help of an external DDI knowledge graph, the decision support
system inputs patient features and outputs medication sug-
gestions and the corresponding DDI explanations to doctors
as clinical diagnostic references. The design of DSSDDI is
depicted in Fig. 1.
There are three modules in DSSDDI: the Drug-Drug Inter-
action (DDI) module, the Medical Decision (MD) module and
the Medical Support (MS) module. The function and merit of
each module are described as follows.
• The DDI module uses our proposed Drug-Drug Interac-
tion Graph Convolutional Network (DDIGCN) to learn
drug representations from synergistic or antagonistic ef-
fects between drugs. The DDI module can alleviate severe
adverse drug events, which is crucial to ensure safe and
effective medical decisions.
• The MD module uses a Medical Decision Graph Con-
volutional Network (MDGCN) to suggest drugs. We
arXiv:2303.02405v1 [cs.LG] 4 Mar 2023
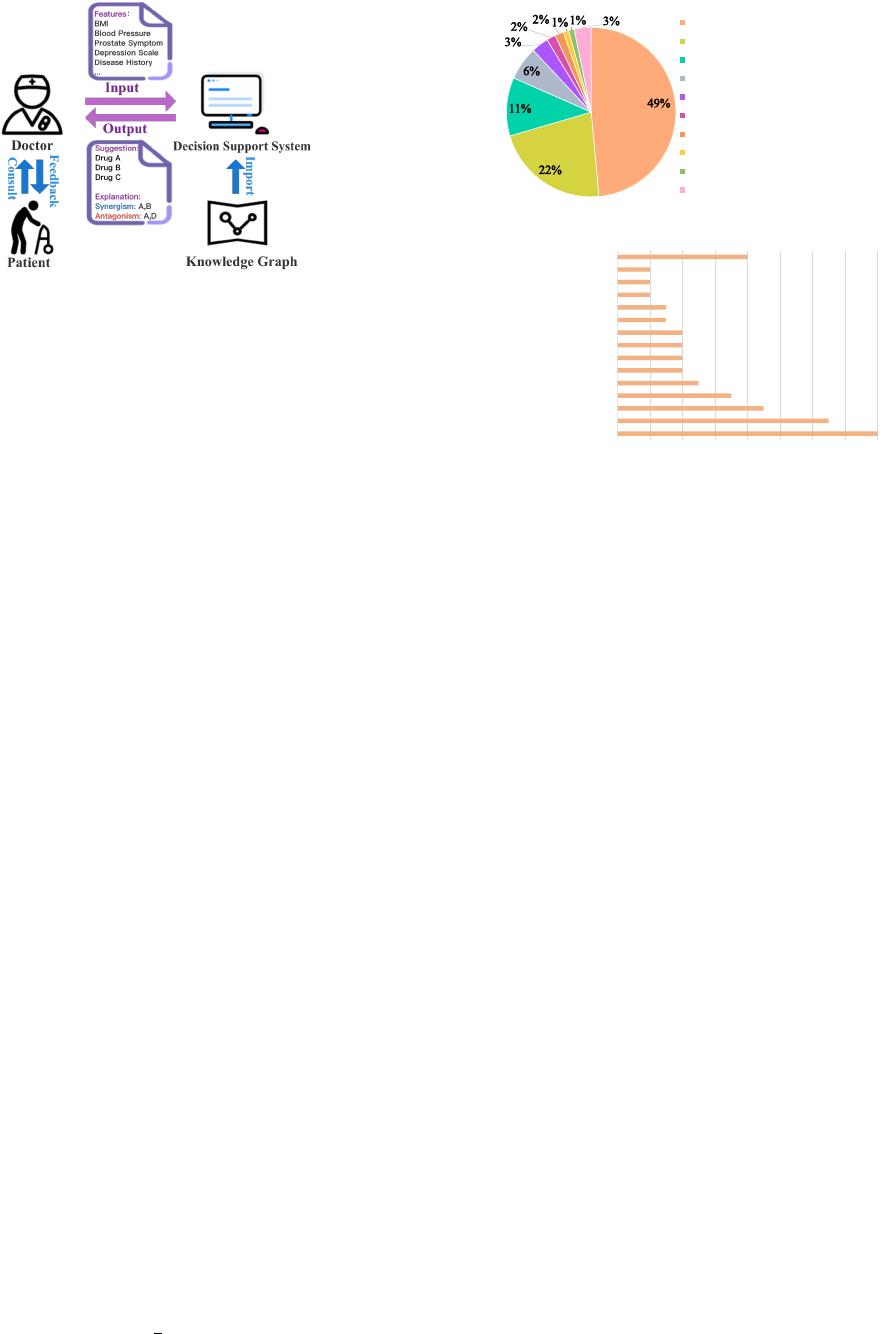
Fig. 1. Our proposed decision support system uses external knowledge of
DDI. Given patient features as input, a doctor can obtain the medication
suggestions from the system, as well as the corresponding DDI explanations.
construct counterfactual links to augment the training data
for MDGCN based on the causal model that considers the
representations of patients and drugs as context, DDI and
patients’ similarity as treatment, and medication use as
outcome. This can learn the causal relationship between
DDI and medication use.
• After obtaining the suggested drugs, the MS module
extracts coherent subgraphs with DDI knowledge as
explainable factors for doctors’ reference. Such subgraphs
illustrate the synergistic and antagonistic effects between
drugs.
Experiments on data from Hong Kong Chronic Disease Study
Project and public diagnostic data MIMIC-III [9] demonstrate
the superior performance of DSSDDI in medication suggestion
and its explainability.
II. DATA COLLECTION
Our study focuses on the patients participating in the Hong
Kong Chronic Disease Study Project, who may require mul-
tiple medications because they have multi-chronic diseases.
We collect data from questionnaire interviews and laboratory
results to predict what medications they would need to take.
Since drug-drug interactions are the primary consideration for
doctors when prescribing medications, DSSDDI is designed to
avoid the inclusion of drugs that contain antagonistic effects
between each other and suggest drugs that have synergistic
effects. In this section, we first introduce the participants
enrolled in this project, then describe the drugs we use for
the decision support system, followed by a further description
of the drug-drug interactions used in this paper.
A. Participants
This project was initiated by Prince of Wales Hospital
1
in 2001. Subjects aged 65 years and older were recruited
under the project. The cohort was invited for a questionnaire
interview and measurement of physical performance for 1 – 4
times during 2001 – 2017. We extracted 2254 male and 1903
female interview records. The distribution of the diseases
suffered by these subjects is shown in Fig. 2. Hypertension,
1
https://www3.ha.org.hk/pwh/index e.asp
Hypertension (49%)
Cardiovascular Events (22%)
Type 2 Diabetes Mellitus (11%)
Gastric or Duodenal Ulcer (6%)
Arthritis (3%)
Prostatic Hyperplasia (2%)
Diabetic Nephropathy (2%)
Myocardial Infarction (1%)
Asthma (1%)
Other Diseases (3%)
Fig. 2. The proportion of patients with various diseases.
0 2 4 6 8 10 12 14 16
Hypertension
Cardiovascular Events
Arthritis
Erosive Esophagitis
Type 2 Diabetes Mellitus
Diabetic Nephropathy
Seizures
Gastric or Duodenal Ulcer
Eye Diseases
Anxiety Disorder
Edema
Prostatic Hyperplasia
Asthma
Thromboembolism
Other Diseases
The Number of Medications
Fig. 3. The distribution of medications for common chronic diseases.
cardiovascular diseases, diabetes, digestive diseases and arthri-
tis are the chronic diseases they commonly suffer.
The questionnaire interview contains three types of subject
information. The first type is the personal information about
the participants such as gender and age. The second type is the
clinical history of participants. For example, participants were
asked whether they had prostatitis before or had taken taken
the Alpha-blocker. It is important to note that the questionnaire
only mentions the clinical history of the family of drugs, but
not the specific drugs. The third type is a psychological assess-
ment of the participants, including the Geriatric Depression
Scale (GDS) Score and some emotional questions, such as
whether they had felt downhearted in the last four weeks. The
physical examination included the participants’ blood pressure,
Body Mass Index (BMI), etc. Combining the three types of
information, we collected a total of 71 features.
B. Medication Use
In total, the participants took 86 medications that are
commonly used to treat chronic conditions. For example,
Doxazosin, a medication commonly used by the participants,
is an alpha-1 adrenergic receptor used to treat mild to moderate
hypertension and urinary obstruction due to benign prostatic
hyperplasia. Fig. 3 shows the distribution of these 86 drugs
for various diseases. As there is usually more than one drug
available for treating a chronic disease such as diabetes,
gastrointestinal diseases, and arthritis, the choice of the most
appropriate drug is a significant consideration for doctors,
and our proposed DSSDDI is designed to help doctors make
decisions more efficiently. We collect pre-trained embedding
of each drug in the Drug Repurposing Knowledge Graph
(DRKG) [10] as the original feature of the drug for the
medication suggestion prediction. Each pre-trained embedding
is trained using a classical knowledge representation learning
method named TransE [11] with a dimension size of 400.

C. Drug-Drug Interactions
DrugCombDB [12] is a database containing drug-drug in-
teractions obtained from various sources, including external
databases, manual curations from PubMed literature and ex-
perimental results. We take the drug-drug interactions that have
been classified as exhibiting synergistic or antagonistic effects
from DrugCombDB
2
. For the 86 drugs used for suggestion,
we extract 97 drug pairs classified as having synergistic effects
and 243 drug pairs classified as having antagonistic effects
from the DrugCombDB database. Based on these drug-drug
interactions, the proposed decision support system obtains
better effectiveness by avoiding pairs with antagonistic effects
and promoting pairs with synergistic effects in medication
suggestions.
III. PROBLEM FORMULATION
In this paper, we design three modules for the proposed
decision support system: Drug-Drug Interaction (DDI) mod-
ule, Medical Decision (MD) module and Medical Support
(MS) module. In this section, we will first introduce the
generalized decision support system, then define the DDI
graph constructed in the DDI module, and give the definitions
of medical decision and medical support.
Definition 1 (Decision Support System): Given a set of drug
candidates denoted as V = {D
1
, D
2
, · · · , D
|V |
}, the decision
support system is designed to suggest a list of appropriate
drugs from V to a patient S
i
based on the patient features x
i
.
Definition 2 (Drug-Drug Interaction (DDI) Graph): We
define the drug-drug interaction (DDI) graph as G = (V, E),
where the node set V = {D
1
, D
2
, · · · , D
|V |
} denotes the
drugs and the edge set E denotes the synergistic or antagonis-
tic effects between drugs. An edge e
uv
= 1 in E represents
a synergistic effect between drugs D
u
and D
v
, and an edge
e
uv
= −1 in E represents an antagonistic effect.
Definition 3 (Medical Decision): The target of medical
decision is to identify the most effective combination of drugs
for the patients from a set of drug candidates. By representing
the relationship between patients and drugs as a bipartite
graph, we formulate the medical decision problem based on
the results of link prediction and further use the drug-drug
interactions as constraints to identify the appropriate drugs.
Specifically, suppose that we are given a set of observed
patients’ data denoted as O = {x
i
, y
i
}
m
i=1
, y
i
is a vector with
y
iv
= 1 if patient S
i
is taking drug D
v
and y
iv
= 0 otherwise,
m is the number of observed patients. The set of unobserved
patients’ data is denoted as U = {x
j
, y
j
}
n
j=m+1
, where n
is the number of all patients. Given the patient features x
j
,
the target of medical decision is first to predict the score
of each drug and then to suggest the most k reliable drugs
Q = {D
q 1
, D
q 2
, · · · , D
q k
} to the unobserved patient S
j
based
on drug-drug interactions.
Definition 4 (Medical Support): The target of medical
support is to find explainable factors through drug-drug inter-
2
http://drugcombdb.denglab.org/download/
actions for the k suggested drugs Q = {D
q 1
, D
q 2
, · · · , D
q k
}.
Specifically, given Q and the DDI graph G, we can find
a subgraph G
sub
of G containing all drug-drug interactions
associated with the suggested drugs, and thus can act as
medical support for the Medical Decision module.
IV. THE PROPOSED DSSDDI
Fig. 4 depicts the overall architecture of DSSDDI which
consists of three modules: Drug-Drug Interaction, Medical
Decision and Medical Support. In Drug-Drug Interaction, we
learn the drug relation representations. In Medical Decision,
we capture the causal relationship between DDI and medi-
cation use. In Medical Support, we generate the explanation
of the suggested drugs. In this section, we elaborate on each
module.
A. Drug-Drug Interaction Module
In the Drug-Drug Interaction (DDI) module, we first de-
velop a model, DDIGCN, to learn the drug representations.
The main idea is to learn drug relation features through
synergistic or antagonistic effects between drugs. In the fol-
lowing, we first describe how to construct the DDI graph, then
illustrate how to update the drug representations by DDIGCN,
and finally describe the model training process.
1) DDI Graph Construction: As described in Definition 2,
we construct the DDI graph G = (V, E) based on the data
collected from DrugCombDB [12] with Drug ID embedding
vectors z
v
for D
v
∈ V . To better capture the relation features
among drugs, we use one-hot ID embeddings instead of
pre-trained embeddings as the original features in this DDI
module. Besides synergistic and antagonistic effects, we add
the third type of edges between drugs in the DDI graph to
explicitly indicate that they have no interactions. Specifically,
we randomly sample drug pairs, denoted as D
u
and D
v
, from
V with no synergistic or antagonistic effect, and create an
edge e
uv
= 0 to represent the lack of interactions between
them. In this manner, DDIGCN can capture synergistic and
antagonistic drug-drug interactions as well as no interactions
in drug embeddings.
2) DDIGCN: In this step, we design a DDIGCN to update
the drug representations. We use Graph Isomorphism Network
(GIN) [13] as the backbone. The graph convolutional operation
is defined as:
z
(t)
v
= f
(t)
Θ
1
(1 +
(t)
) · z
(t−1)
v
+
P
u∈N
v
z
(t−1)
u
|N
v
|
!
, (1)
where z
(t)
v
denotes the updated hidden representation of drug
D
v
after t layers propagation, f
(t)
Θ
1
denotes the multi-layer
perceptrons (MLP) with parameters Θ
1
,
(t)
is a learnable
parameter of the t-th graph convolutional layer, N
v
denotes
the set of drugs that have interactions with drug D
v
.
Besides GIN, we also consider signed graph-based models,
such as SGCN [14], SiGAT [15] and SNEA [16] as alternative
backbones, as there are both positive and negative edges in the
DDI graph G. Take SGCN as an example, we denote B
v
(t) =
{D
u
|e
uv
= 1} and U
v
(t) = {D
u
|e
uv
= −1} for drug D
v
.
of 14
免费下载
【版权声明】本文为墨天轮用户原创内容,转载时必须标注文档的来源(墨天轮),文档链接,文档作者等基本信息,否则作者和墨天轮有权追究责任。如果您发现墨天轮中有涉嫌抄袭或者侵权的内容,欢迎发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。

评论